Temple Run Oz
2013-08-31T23:48:00-07:00
phoenix user
ANDROID|android-games|
kaspersy mobile security
get the cracked version of kaspersky mobile security....absolutely for free...
*you will be up-to-date with the latest virus definitions and you will be secure and protected...
*install this app on your android device...create a free account ...and enjoy kaspersky anti-theft protection for your device... for life time...
(anti-theft protection is super cool of all of its amazing features..)
*you can block unwanted calls,texts with blocking list....
*and many more.....with this tiny smart app.....
kaspersy mobile security
2013-08-26T06:57:00-07:00
phoenix user
ANDROID|
Fundamentals of DataBase Syatems 6thEdition
Fundamentals of DataBase Syatems 6thEdition
2013-08-24T10:05:00-07:00
Unknown
Introduction to Algorithims 3rd edition
Introduction to Algorithims 3rd edition
by: T H O M A S H. C H A R L E S E.
R O N A L D L . C L I F F O R D S T E I N
C O R M E NL E I S E R S O N
R I V E S T

Download full text here : CLICK
by: T H O M A S H. C H A R L E S E.
R O N A L D L . C L I F F O R D S T E I N
C O R M E NL E I S E R S O N
R I V E S T
Download full text here : CLICK
Introduction to Algorithims 3rd edition
2013-08-24T10:02:00-07:00
Unknown
Computer networks -A Tanenbaum 5th edition
Computer networks -A Tanenbaum 5th edition
2013-08-24T09:56:00-07:00
Unknown
Big Data Analytics
Big Data Analytics
How an agile approach helps you quickly realise the value of big data
Don’t get bogged down by Big Data
Big data is massive and messy, and it’s coming at you fast. These characteristics pose a problem for data storage and processing, but focusing on these factors has resulted in a lot navel-gazing and an unnecessary emphasis on technology.
It’s not about Data. It’s about Insight and Impact
The potential of Big Data is in its ability to solve business problems and provide new business opportunities. So to get the most from your Big Data investments, focus on the questions you’d love to answer for your business. This simple shift can transform your perspective, changing big data from a technological problem to a business solution.


What would you really like to know about your business?
- Why are our customers leaving us?
- What is the value of a 'tweet' or a 'like'?
- What products are our customers most likely to buy?
- What is the best way to communicate with our customers?
- Are our investments in customer service paying off?
- What is the optimal price for my product right now?
The value of data is only realised through insight. And insight is useless until it’s turned into action. To strike upon insight, you first need to know where to dig. Finding the right questions will lead you to the well.
2. Accelerate Insight
The facts you need are likely buried within a jumble of legacy systems, hidden in plain sight. Big Data can uncover those facts, but typical analytics projects can turn into expensive, time-consuming technology efforts in search of problems to solve.
We offer a different way.
Agile analytics allow you to achieve big results, little by little, by encompassing:
- Big data technologies: select the tools and techniques which will best handle the data for your specific goals, timeframe and architecture.
- Continuous delivery: deliver business value early and often. Build your platform over time, not all up front.
- Advanced analytics: apply sophisticated statistical techniques to extract actionable knowledge and insights.
3. Learn and apply
Lean learning provides actionable results more rapidly
Our lean learning approach measures the value you gain at each step, in small iterations. Use the results to improve the process or change course to a more fruitful direction.
The benefits of an agile approach:
- Realise the value from your data much faster than other methods.
- Save money by avoiding the waste inherent in building your platform before answering crucial business questions.
- Free your team to pivot when unexpected insights emerge.

Big Data Analytics
2013-08-24T09:43:00-07:00
Unknown
Time-series forecasting: Bike Accidents
Time-series forecasting: Bike Accidents
About a year ago I posted this video visualization of all the reported accidents involving bicycles in Montreal between 2006 and 2010. In the process I also calculated and plotted the accident rate using a monthly moving average. The results followed a pattern that was for the most part to be expected. The rate shoots up in the spring, and declines to only a handful during the winter months.
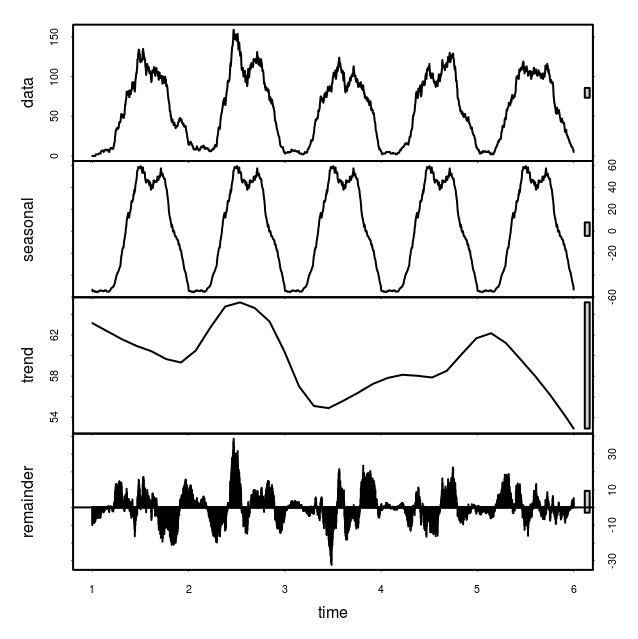
It’s now 2013 and unfortunately our data ends in 2010. However, the pattern does seem to be quite regular (that is, exhibits annual periodicity) so I decided to have a go at forecasting the time series for the missing years. I used a seasonal decomposition of time series by LOESS to accomplish this.
You can see the code on github but here are the results. First, I looked at the four components of the decomposition:
Indeed the seasonal component is quite regular and does contain the intriguing dip in the middle of the summer that I mentioned in the first post.
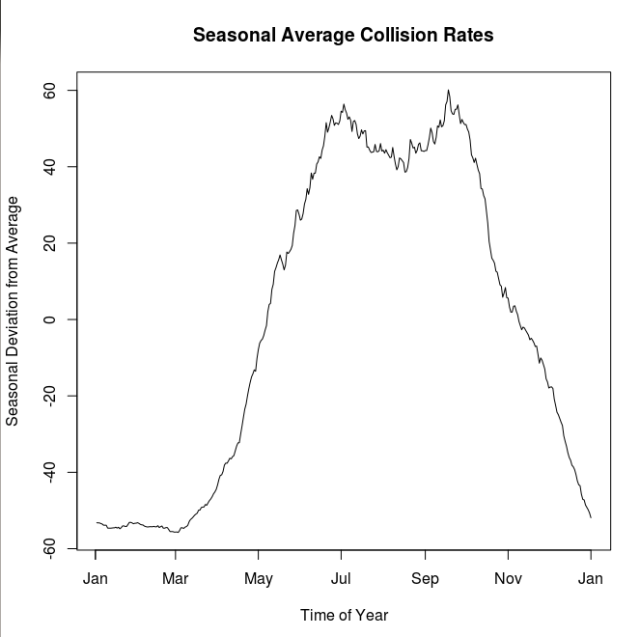
This figure shows just the seasonal deviation from the average rates. The peaks seem to be early July and again in late September. Before doing any seasonal aggregation I thought that the mid-summer dip may correspond with the mid-August construction holiday, however it looks now like it is a broader summer-long reprieve. It could be a population wide vacation effect.
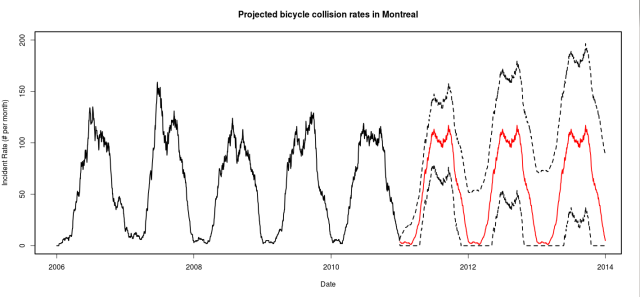
Finally, I used an exponential smoothing model to project the accident rates into the 2011-2013 seasons.
It would be great to get the data from these years to validate the forecast, but for now lets just hope that we’re not pushing up against those upper confidence bounds.
Time-series forecasting: Bike Accidents
2013-08-24T09:33:00-07:00
Unknown
Design of Experiments: General Background
Design of Experiments: General Background
June 2nd, 2013
The statistical methodology of design of experiments has a long history starting back with the work of Fisher, Yates and other researchers. One of the main motivating factors is to make gooduse of available resources and to avoid making decisions that cannot be corrected during the analysis stage of an investigation.
The statistical methodology is based on a systematic approach to investigate the causes of variation of a system of interest and to control the factors that can be while taking some account of nuisance factors that can be measured but not controlled by an experimenter. As with most things there are some general principles and common considerations for experiments run in a variety of different areas.
The following considerations are required for running an experiment:
- Absence of Systematic Error: when running an experiment the aim is to obtain a correctestimate of the metric of interest, e.g. treatment effect or difference. The design selected should avoid the introduction of bias into the subsequent analysis.
- Adequate Precision: an experimental design is chosen to allow estimation and comparison of effects of interest, e.g. differences between treatments, so there should be sufficient replication in the experiment to allow these effects to be precisely estimated and also for meaningful differences to be detected.
- Range of Validity: the range of variables considered in the experiment so cover the range of interest so that the results can be generalised without need to rely on extrapolation.
- Simplicity: ideally the choice of design should be simple to implement to ensure that it can be run as intended and to reduce the chance of missing data which could impact on the analysis of the results.
There are various objectives from using design of experiments methodology and these include (a) screening of a large number of factors to reduce this set to a more manageable subset that can be investigated in greater detail, (b) response surface methodology to understand the behaviour or a system and (c) optimisation of a process or reduction of uncontrollable variation (or noise) in a process.
There are not a large number of packages relating to design of experiments in R but those of interest are covered by the Task View on CRAN.
Design of Experiments: General Background
2013-08-24T09:29:00-07:00
Unknown
Fitting a Model by Maximum Likelihood
Fitting a Model by Maximum Likelihood
Maximum Likehood Estimation (MLE) is a statistical technique for estimating model parameters. It basically sets out to answer the question: what model parameters are most likely to characterise a given set of data? First you need to select a model for the data. And the model must have one or more (unknown) parameters. As the name implies, MLE proceeds to maximise a likelihood function, which in turn maximises the agreement between the model and the data.
Most illustrative examples of MLE aim to derive the parameters for a probability density function (PDF) of a particular distribution. In this case the likelihood function is obtained by considering the PDF not as a function of the sample variable, but as a function of distribution’s parameters. For each data point one then has a function of the distribution’s parameters. The joint likelihood of the full data set is the product of these functions. This product is generally very small indeed, so the likelihood function is normally replaced by a log-likelihood function. Maximising either the likelihood or log-likelihood function yields the same results, but the latter is just a little more tractable!
Fitting a Normal Distribution
Let’s illustrate with a simple example: fitting a normal distribution. First we generate some data.
> set.seed(1001)>> N <- 100>> x <- rnorm(N, mean = 3, sd = 2)>> mean(x)[1] 2.998305> sd(x)[1] 2.288979 |
Then we formulate the log-likelihood function.
> LL <- function(mu, sigma) {+ R = dnorm(x, mu, sigma)+ #+ -sum(log(R))+ } |
And apply MLE to estimate the two parameters (mean and standard deviation) for which the normal distribution best describes the data.
> library(stats4)>> mle(LL, start = list(mu = 1, sigma=1))Call:mle(minuslogl = LL, start = list(mu = 1, sigma = 1))Coefficients: mu sigma2.998305 2.277506Warning messages:1: In dnorm(x, mu, sigma) : NaNs produced2: In dnorm(x, mu, sigma) : NaNs produced3: In dnorm(x, mu, sigma) : NaNs produced |
Those warnings are a little disconcerting! They are produced when negative values are attempted for the standard deviation.
> dnorm(x, 1, -1) [1] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN [30] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN [59] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN [88] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN |
There are two ways to sort this out. The first is to apply constraints on the parameters. The mean does not require a constraint but we insist that the standard deviation is positive.
> mle(LL, start = list(mu = 1, sigma=1), method = "L-BFGS-B", lower = c(-Inf, 0), upper = c(Inf, Inf))Call:mle(minuslogl = LL, start = list(mu = 1, sigma = 1), method = "L-BFGS-B", lower = c(-Inf, 0), upper = c(Inf, Inf))Coefficients: mu sigma2.998304 2.277506 |
This works because mle() calls optim(), which has a number of optimisation methods. The default method is BFGS. An alternative, the L-BFGS-B method, allows box constraints.
The other solution is to simply ignore the warnings. It’s neater and produces the same results.
> LL <- function(mu, sigma) {+ R = suppressWarnings(dnorm(x, mu, sigma))+ #+ -sum(log(R))+ }>> mle(LL, start = list(mu = 1, sigma=1))Call:mle(minuslogl = LL, start = list(mu = 1, sigma = 1))Coefficients: mu sigma2.998305 2.277506 |
The maximum-likelihood values for the mean and standard deviation are damn close to the corresponding sample statistics for the data. Of course, they do not agree perfectly with the values used when we generated the data: the results can only be as good as the data. If there were more samples then the results would be closer to these ideal values.
A note of caution: if your initial guess for the parameters is too far off then things can go seriously wrong!
> mle(LL, start = list(mu = 0, sigma=1))Call:mle(minuslogl = LL, start = list(mu = 0, sigma = 1))Coefficients: mu sigma 51.4840 226.8299 |
Fitting a Linear Model
Now let’s try something a little more sophisticated: fitting a linear model. As before, we generate some data.
> x <- runif(N)> y <- 5 * x + 3 + rnorm(N) |
We can immediately fit this model using least squares regression.
> fit <- lm(y ~ x)>> summary(fit)Call:lm(formula = y ~ x)Residuals: Min 1Q Median 3Q Max-1.96206 -0.59016 -0.00166 0.51813 2.43778Coefficients: Estimate Std. Error t value Pr(>|t|)(Intercept) 3.1080 0.1695 18.34 <2e-16 ***x 4.9516 0.2962 16.72 <2e-16 ***---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.8871 on 98 degrees of freedomMultiple R-squared: 0.7404, Adjusted R-squared: 0.7378F-statistic: 279.5 on 1 and 98 DF, p-value: < 2.2e-16 |
The values for the slope and intercept are very satisfactory. No arguments there. We can superimpose the fitted line onto a scatter plot.
> plot(x, y)> abline(fit, col = "red") |
Pushing on to the MLE for the linear model parameters. First we need a likelihood function. The model is not a PDF, so we can’t proceed in precisely the same way that we did with the normal distribution. However, if you fit a linear model then you want the residuals to be normally distributed. So the likelihood function fits a normal distribution to the residuals.
LL <- function(beta0, beta1, mu, sigma) { # Find residuals # R = y - x * beta1 - beta0 # # Calculate the likelihood for the residuals (with mu and sigma as parameters) # R = suppressWarnings(dnorm(R, mu, sigma)) # # Sum the log likelihoods for all of the data points # -sum(log(R))} |
One small refinement that one might make is to move the logarithm into the call to dnorm().
LL <- function(beta0, beta1, mu, sigma) { R = y - x * beta1 - beta0 # R = suppressWarnings(dnorm(R, mu, sigma, log = TRUE)) # -sum(R)} |
It turns out that the initial guess is again rather important and a poor choice can result in errors. We will return to this issue a little later.
> fit <- mle(LL, start = list(beta0 = 3, beta1 = 1, mu = 0, sigma=1))Error in solve.default(oout$hessian) : system is computationally singular: reciprocal condition number = 3.01825e-22> fit <- mle(LL, start = list(beta0 = 5, beta1 = 3, mu = 0, sigma=1))Error in solve.default(oout$hessian) : Lapack routine dgesv: system is exactly singular: U[4,4] = 0 |
But if we choose values that are reasonably close then we get a decent outcome.
> fit <- mle(LL, start = list(beta0 = 4, beta1 = 2, mu = 0, sigma=1))> fitCall:mle(minuslogl = LL, start = list(beta0 = 4, beta1 = 2, mu = 0, sigma = 1))Coefficients: beta0 beta1 mu sigma 3.5540217 4.9516133 -0.4459783 0.8782272 |
The maximum-likelihood estimates for the slope (beta1) and intercept (beta0) are not too bad. But there is a troubling warning about NANs being produced in the summary output below.
> summary(fit)Maximum likelihood estimationCall:mle(minuslogl = LL, start = list(beta0 = 4, beta1 = 2, mu = 0, sigma = 1))Coefficients: Estimate Std. Errorbeta0 3.5540217 NaNbeta1 4.9516133 0.2931924mu -0.4459783 NaNsigma 0.8782272 0.0620997-2 log L: 257.8177Warning message:In sqrt(diag(object@vcov)) : NaNs produced |
It stands to reason that we actually want to have the zero mean for the residuals. We can apply this constraint by specifying mu as a fixed parameter. Another option would be to simply replace mu with 0 in the call to dnorm(), but the alternative is just a little more flexible.
> fit <- mle(LL, start = list(beta0 = 2, beta1 = 1.5, sigma=1), fixed = list(mu = 0), nobs = length(y))> summary(fit)Maximum likelihood estimationCall:mle(minuslogl = LL, start = list(beta0 = 2, beta1 = 1.5, sigma = 1), fixed = list(mu = 0), nobs = length(y))Coefficients: Estimate Std. Errorbeta0 3.1080423 0.16779428beta1 4.9516164 0.29319233sigma 0.8782272 0.06209969-2 log L: 257.8177 |
The resulting estimates for the slope and intercept are rather good. And we have standard errors for these parameters as well.
How about assessing the overall quality of the model? We can look at the Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC). These can be used to compare the performance of different models for a given set of data.
> AIC(fit)[1] 263.8177> BIC(fit)[1] 271.6332> logLik(fit)'log Lik.' -128.9088 (df=3) |
Returning now to the errors mentioned above. Both of the cases where the call to mle() failed resulted from problems with inverting the Hessian Matrix. With the implementation of mle() in the stats4 package there is really no way to get around this problem apart from having a good initial guess. In some situations though, this is just not feasible. There are, however, alternative implementations of MLE which circumvent this problem. The bbmle package has mle2() which offers essentially the same functionality but includes the option of not inverting the Hessian Matrix.
> library(bbmle)> > fit <- mle2(LL, start = list(beta0 = 3, beta1 = 1, mu = 0, sigma = 1))> > summary(fit)Maximum likelihood estimationCall:mle2(minuslogl = LL, start = list(beta0 = 3, beta1 = 1, mu = 0, sigma = 1))Coefficients: Estimate Std. Error z value Pr(z) beta0 3.054021 0.083897 36.4019 <2e-16 ***beta1 4.951617 0.293193 16.8886 <2e-16 ***mu 0.054021 0.083897 0.6439 0.5196 sigma 0.878228 0.062100 14.1421 <2e-16 ***---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1-2 log L: 257.8177 |
Here mle2() is called with the same initial guess that broke mle(), but it works fine. The summary information for the optimal set of parameters is also more extensive.
Fitting a linear model is just a toy example. However, Maximum-Likelihood Estimation can be applied to models of arbitrary complexity. If the model residuals are expected to be normally distributed then a log-likelihood function based on the one above can be used. If the residuals conform to a different distribution then the appropriate density function should be used instead of dnorm().
Fitting a Model by Maximum Likelihood
2013-08-24T09:28:00-07:00
Unknown
Subscribe to:
Posts (Atom)
referal
http://kisscartoon.me/G/169319?l=http%3a%2f%2fkisscartoon.me%2f
Subscribe me
Space for adds
Blog Archive
-
▼
2013
(161)
-
▼
Aug
(15)
- Temple Run Oz
- kaspersy mobile security
- Fundamentals of DataBase Syatems 6thEdition
- Introduction to Algorithims 3rd edition
- Computer networks -A Tanenbaum 5th edition
- Big Data Analytics
- Time-series forecasting: Bike Accidents
- Design of Experiments: General Background
- Fitting a Model by Maximum Likelihood
- Step by step to build my first R Hadoop System
- Big Data Sets you can use with R
- What are the hottest areas for CS Research? (Based...
- R vs Python Speed Comparison for Bootstrapping
- TINTIN COMICS COMPLETE COLLECTION FREE DOWNLOAD
- AIRCEL 3G Mornings Unlimited 3g Offer in INDIA
-
▼
Aug
(15)
Popular Posts
-
 TINTIN COMICS COMPLETE COLLECTION FREE DOWNLOAD
TINTIN COMICS COMPLETE COLLECTION FREE DOWNLOAD
-
 ILEAP FULL SOFTWARE DOWNLOAD
ILEAP FULL SOFTWARE DOWNLOAD
-
 SIDNEY SHELDON COMPLETE COLLECTION FREE DOWNLOAD
SIDNEY SHELDON COMPLETE COLLECTION FREE DOWNLOAD
-
 MICHAEL CHRICTON NOVELS COMPLETE COLLECTION FREE DOWNLOAD
MICHAEL CHRICTON NOVELS COMPLETE COLLECTION FREE DOWNLOAD
-
 HARRY POTTER COMPLETE COLLECTION DOWNLOAD
HARRY POTTER COMPLETE COLLECTION DOWNLOAD
-
 ASTERIX AND OBELIX COMICS COMPLETE COLLECTION
ASTERIX AND OBELIX COMICS COMPLETE COLLECTION
-
 AZHAGI INDIAN LANGUAGE WORD PROCESSOR
AZHAGI INDIAN LANGUAGE WORD PROCESSOR
-
 xiaomi mi3 : the cheapest mobile with latest features
xiaomi mi3 : the cheapest mobile with latest features
-
 1 NENOKKADINE FULL SONGS MP3 DOWNLOAD
1 NENOKKADINE FULL SONGS MP3 DOWNLOAD
Labels
A - TELUGU
(1)
A.R.RAHMAN
(12)
ANDROID
(18)
android-games
(12)
B - TELUGU
(4)
COMICS
(3)
Devi Sri Prasad
(5)
ENGLISH NOVELS
(9)
EXPERIA n EXPRESSO
(12)
J - TELUGU
(1)
K - TELUGU
(1)
M - TELUGU
(1)
N - TELUGU
(1)
PHOENIX MUSIC
(26)
S.S.Thaman
(4)
Sandeep Chowtha
(1)
Self-help Literature
(1)
SIDDARTH
(1)
SOFT-FACT
(50)
SOFTWARES
(23)
TELUGU NOVELS
(10)
TELUGU SONGS
(26)
V - Telugu
(1)
Banner2